ScripyTCR术语表
克隆 (Clone) 和克隆型 (Clonotype): 在免疫学中,特别是T细胞或B细胞的语境下,一个“克隆”指的是一群起源于同一个祖细胞的细胞。这些细胞因为共享相同的T细胞受体 (TCR) 或B细胞受体 (BCR) 而被定义为一个“克隆型”。这意味着它们能够识别和响应同一种特定的抗原。
扩增 (Expansion): 当一个T细胞(或B细胞)遇到它能够识别的特定抗原(比如来自病原体或肿瘤细胞的抗原)并被激活时,它会开始快速分裂和增殖,产生大量具有相同TCR(即属于同一克隆型)的子细胞。这个过程就叫做“克隆扩增”。
克隆型大小 (Clonotype Size): 就是每个克隆型中包含的细胞数量。一个克隆型的细胞数量越多,说明这个克隆型发生的“扩增”程度越高。
AIRR
Adaptive Immune Receptor Repertoire.
Within the Scirpy documentation, we simply speak of immune receptors (IR).
The AIRR community defines standards around AIRR data. Scirpy supports the AIRR Rearrangement schema and complies with the AIRR Software Guidelines.
Alellically included B-cells
A B cell with two pairs of IG chains.
See Dual IR.
awkward array
Awkward arrays are a data structure that allows representing nested, variable sized data (such as lists of lists, lists of dictionaries).
It is computationally efficient and can be manipulated with NumPy-like idioms.
For more details, check out the awkward documentation.
BCR
B-cell receptor.
A BCR consists of two Immunoglobulin (IG) heavy chains and two IG light chains.
The two light chains contain a variable region, which is responsible for antigen recognition.
BCR 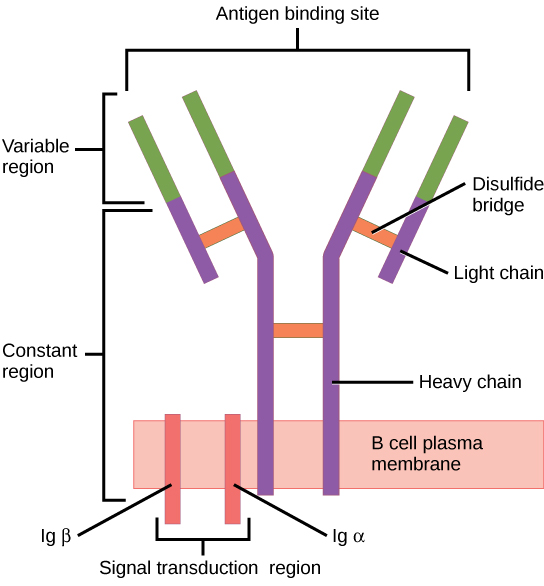
Image By CNX OpenStax under the CC BY-4.0 license, obtained from Wikimedia Commons
CDR
Complementary-determining region.
The diversity and, therefore, antigen-specificity of IRs is predominantly determined by three hypervariable loops (CDR1, CDR2, and CDR3) on each of the α- and β receptor arms.
CDR1 and CDR2 are fully encoded in germline V genes. In contrast, the CDR3 loops are assembled from V, (D), and J segments and comprise random additions and deletions at the junction sites.
Thus, CDR3 regions make up a large part of the adaptive immune receptor variability and are therefore thought to be particularly important for antigen specificity (reviewed in [AHS15]).
CDR
Image from [AHS15] under the CC BY-NC-SA-3.0 license.
CDR3
Complementary-determining region 3.
See CDR.
Chain locus
Scirpy supports all valid IMGT locus names:
Loci with a VJ junction
- TRA (T-cell receptor alpha)
- TRG (T-cell receptor gamma)
- IGL (Immunoglobulin lambda)
- IGK (Immunoglobulin kappa)
Loci with a VDJ junction
- TRB (T-cell receptor beta)
- TRD (T cell receptor delta)
- IGH (Immunoglobulin heavy chain)
Clonotype
A clonotype designates a collection of T or B cells that descend from a common, antecedent cell, and therefore, bear the same adaptive immune receptors and recognize the same epitopes.
In single-cell RNA-sequencing (scRNA-seq) data, T or B cells sharing identical complementarity-determining regions 3 (CDR3) nucleotide sequences of both VJ and VDJ chains (e.g. both α and β TCR chains) make up a clonotype.
Scirpy provides a flexible approach to clonotype definition based on CDR3 sequence identity or similarity. Additionally, it is possible to require clonotypes to have the same V-gene, enforcing the CDR 1 and 2 regions to be the same.
For more details, see the page about our IR model and the API documentation of scirpy.tl.define_clonotypes().
Clonotype cluster
A higher-order aggregation of clonotypes that have different CDR3 nucleotide sequences, but might recognize the same antigen because they have the same or similar CDR3 amino acid sequence.
This is especially relevant for BCR, because clonally related cells are likely to differ due to somatic hypermutation.
It is important to understand that there is currently no best practice or go-to approach on how to define clonotype clusters for BCR, as it remains an active research field ([YK15]).
There exist many different approaches such as maximum-likelihood ([RM16]), hierarchical clustering ([GAB+17]), spectral clustering ([NK18]), natural language processing ([LNKK21]) and network based approaches ([BRPH+13]).
A recent comparison study indicates that computationally more sophisticated clonal inference approaches do not outperform simplistic, computationally cheaper ones ([BvanSchaikS+24]). That said, there is still a need for more in-depth comparison studies to confirm these results.
See also: scirpy.tl.define_clonotype_clusters().
Clonotype modularity
The clonotype modularity measures how densely connected the transcriptomics neighborhood graph underlying the cells in a clonotype is.
Clonotypes with a high modularity consist of cells that are transcriptionally more similar than that of a clonotype with a low modularity.
See also scirpy.tl.clonotype_modularity()
Convergent evolution of clonotypes
It has been proposed that IRs are subject to convergent evolution, i.e. a selection pressure that leads to IRs recognizing the same antigen ([VKP+06]).
Evidence of convergent evolution could be clonotypes with the same CDR3 amino acid sequence, but different CDR3 nucleotide sequences (due to synonymous codons) or clonotypes with highly similar CDR3 amino acid sequences that recognize the same antigen.
Dual IR
IRs with more than one pair of VJ and VDJ sequences.
While this was previously thought to be impossible due to the mechanism of allelic exclusion ([BSB10]), there is an increasing amount of evidence for a bona fide dual-IR population ([SB19], [SZY+19], [RobertaPelanda14], [JPG10], [VS10]).
Recent evidence suggests that also B cells with three or more productively rearranged H and/or L chains exist ([ZPWY23]), which indicates how much of B cell development is still unclear.
For more information on how Scirpy handles dual IRs, see the page about our IR model.
Dual TCR
TCRs with more than one pair of α- and β (or γ- and δ) chains.
See Dual IR.
Epitope
The part of an antigen that is recognized by the TCR, BCR, or antibody.
IG
Immunoglobulin
IR
Immune receptor.
Isotypically included B-cells
Similar to Alellically included B-cells, but expresses both IGL and IGK and thus rearrangements are not on alleles of the same gene (= isotypic inclusion).
Multi-tissue clonotype
A clonotype that occurs in multiple tissues of the same patient.
Multichain-cell
Cells with more than two pairs of VJ and VDJ sequences that do not fit into the Dual IR model.
These are usually rare and could be explained by doublets/multiplets, i.e. two or more cells that were captured in the same droplet.
Multichain-cell
(a) UMAP plot of 96,000 cells from [WMdA+20] with at least one detected CDR3 sequence with multichain-cells (n=474) highlighted in green.
(b) Comparison of detected reads per cell in multichain-cells and other cells.
Multichain cells comprised significantly more reads per cell (p = 9.45 × 10⁻²⁵¹, Wilcoxon-Mann-Whitney-test), supporting the hypothesis that (most of) multichain cells are technical artifacts arising from cell-multiplets ([IKK+16]).
Orphan chain
An IR chain is called orphan if its corresponding counterpart has not been detected.
For instance, if a cell has only a VJ chain (e.g. TCR-alpha), but no VDJ chain (e.g. TCR-beta), the cell will be flagged as “Orphan VJ”.
Orphan chains are most likely the effect of stochastic dropouts due to sequencing inefficiencies.
See also scirpy.tl.chain_qc().
Private clonotype
A clonotype that is specific for a certain patient.
Productive chain
Productive chains are IR chains with a CDR3 sequence that produces a functional peptide.
Scirpy relies on the preprocessing tools (e.g. CellRanger or TraCeR) for flagging non-productive chains.
Typically chains are flagged as non-productive if they contain a stop codon or are not within the reading frame.
Public clonotype
A clonotype that is shared across multiple patients, e.g. a clonotype recognizing a common viral epitope.
Public vs Private Clonotype
Image from [SMR+18] under the CC BY-4.0 license.
Receptor subtype
More fine-grained classification of the receptor type into
- α/β T cells
- γ/δ T cells
- IG-heavy/IG-κ B cells
- IG-heavy/IG-λ B cells
See also scirpy.tl.chain_qc().
Receptor type
Classification of immune receptors into BCR and TCR.
See also scirpy.tl.chain_qc().
SHM
Common abbreviation for “Somatic hypermutation”.
This process is unique to BCR and occurs as part of affinity maturation upon antigen encounter.
This process further increases the diversity of the variable domain of the BCR and selects for cells with higher affinity.
SHM introduces around one point mutation per 1000 base pairs ([KLS03]) and is able to introduce (although rare) deletions and/or insertions ([WdBL+98]).
Furthermore, SHM is not a stochastic process, but biased in multiple ways (e.g. intrinsic hot-spot motifs (reviewed in [SD18])).
TCR
T-cell receptor.
A TCR consists of one α and one β chain (or, alternatively, one γ and one δ chain).
Each chain consists of a constant and a variable region.
The variable region is responsible for antigen recognition, mediated by CDR regions.
For more information on how Scirpy represents TCRs, see the page about our receptor model.
TCR
Image from Wikimedia Commons under the CC BY-3.0 license.
Tissue-specific clonotype
A clonotype that only occurs in a certain tissue of a certain patient.
UMI
Unique molecular identifier.
Some single-cell RNA-seq protocols label each RNA with a unique barcode prior to PCR-amplification to mitigate PCR bias.
With these protocols, UMI-counts replace the read-counts generally used with RNA-seq.
V(D)J
The variability of IR chain sequences originates from the genetic recombination of Variable, Diversity and Joining gene segments.
The TCR-α, TCR-ɣ, IG-κ, and IG-λ chains get assembled from V and J loci only.
We refer to these chains as VJ chains in Scirpy.
The TCR-β, TCR-δ, and IG-heavy chains get assembled from all three segments.
We refer to these chains as VDJ-chains in Scirpy.
As an example, the figure below shows how a TCR-α chain is assembled from the tra locus.
V to J recombination joins one of many TRAV segments to one of many TRAJ segments.
Next, introns are spliced out, resulting in a TCR-α chain transcript with V, J and C segments directly next to each other (reviewed in [AHS15]).
V(D)J
Image from [AHS15] under the CC BY-NC-SA-3.0 license.
By Gregor Sturm, Tamas Szabo
© Copyright 2025, Gregor Sturm, Tamas Szabo.